In the hunt for improved antibiotics, it is important to realize that natural products have been, without question, the most prolific source of all medicines. With the advent of massively parallel DNA sequencing, it has become apparent that our knowledge of natural product structure and function is astonishingly incomplete. Exploration of uncharted natural product chemical space will undoubtedly lead to improved, and entirely new, medicines. Thus, our group focuses on elucidating the biosynthesis, structure, and function of natural products. Our primary focus has been on thiazole/oxazole-containing peptides. Characterized examples have activities ranging from antibacterials to virulence-promoting toxins. Therefore, the study of these peptidic natural products allows us to not only better understand bacterial virulence (where pharmacological intervention would constitute a pathogen-specific approach to bacterial infection) but also explore unique chemical architectures, ideally positioning us to introduce new structural classes of antibiotics. Our recent successes and focal areas are highlighted below by research project.
reactivity based screening
We are using small-molecule probes to identify natural products with various organic functional groups
recent publications
- Maxson, T., Tietz, et al., "Targeting reactive carbonyls for identifying natural products and their biosynthetic origins." J. Am. Chem. Soc., (2016). doi:10.1021/jacs.6b06848
- Molloy, E.M., et al., "Biological characterization of the hygrobafilomycin antibiotic JBIR-100 and bioinformatic insights into the hygrolide family of natural products." Bioorg. Med. Chem., (2016). doi:10.1016/j.bmc.2016.05.021
- Cox, C.L., et al., "Nucleophilic 1,4-additions for natural product discovery" ACS Chem. Biol. (2014). doi:10.1021/cb500324n
Genomic sequencing has revealed that Actinobacteria, a historically rich source of FDA-approved drugs, possess far greater natural product biosynthetic potential than previously imagined. One of the challenges associated with discovering novel molecules produced by these unexplored pathways involves determining whether the molecule is being made by the host organism and if the molecule is chemically novel in a process known as dereplication. Rapid determination of chemical novelty is especially of interest since rediscovery of known compounds has led to diminishing returns on investment for the pharmaceutical industry. Our lab has developed a reactivity-based screening (RBS) strategy to circumvent these long-standing challenges. By taking advantage of chemoselective chemistry, we are developing probes to react selectively with functional groups found on various classes of RiPP, polyketide, and non-ribosomal peptide natural products.
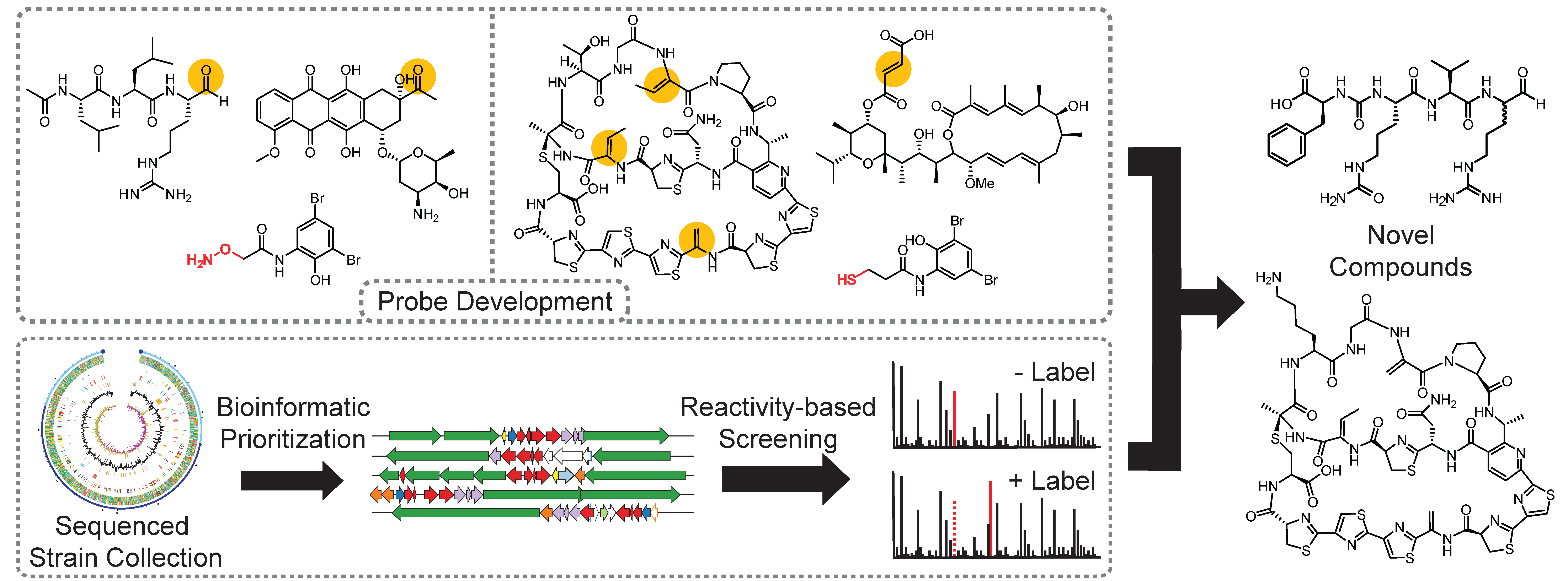
Upon analysis of differential mass spectrometry data and through database searching, we can rapidly determine the presence and novelty of our target natural products. We have taken both a bioinformatics-guided as well as comprehensive screening approach with RBS and have applied this strategy towards the discovery of thiopeptides, fumarate-containing polyketides as well as aldehyde-containing non-ribosomal peptides. Currently, third-generation reporter probes are being developed to target additional organic functional groups with moieties to enhance mass spectrometry-based recognition.